In-silico Analysis of Single Nucleotide Polymorphisms (SNPs) in Human Pten Gene
Abstract:
Phosphatase Tensin Homolog deleted
on Chromosome 10 is a tumour suppressor gene frequently inactivated in human
cancers. Single Nucleotide Polymorphisms (SNPs) are the substitution of one DNA
nucleotide base with another and a commonly occurring genomic alteration. This
study involved analyzing various missense SNPs of the PTEN gene using five
different bioinformatics tools -SIFT, POLYPHEN, CADD, META LR, and MUTATION
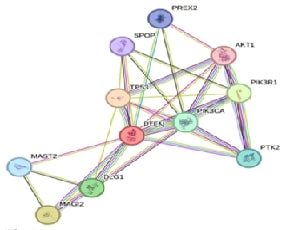
ASSESSOR to identify the tolerated from intolerant ones. String analysis of
PTEN protein-protein interactions was also done using the STRING database. A
total of 8298 missense SNPs were retrieved and analyzed using the five
bioinformatics tools. Out of 8298 missense SNPs analyzed, SIFT categorized 5281
SNPs as deleterious, while POLYPHEN identified 4490 as damaging. CADD showed
that 909 were disease-causing SNPs. META LR identified 2995 as damaging, and the
MUTATION ASSESSOR identified 897 high-risk missense SNPs. This study shows that
the various in silico tools are a good preliminary approach to identifying the
harmful missense SNPs.
References:
[1]. Garcia,
F. A., de O., de Andrade, E. S., & Palmero, E. I., 2022, Insights on
variant analysis in silico tools for pathogenicity prediction. Frontiers
in Genetics, 13, 1010327. https://doi.org/10.3389/fgene.2022.1010327
[2]. Weng,
L. P., Smith, W. M., Dahia, P. L., Ziebold, U., Gil, E., Lees, J. A., &
Eng, C., 1999, PTEN suppresses breast cancer cell growth by phosphatase
activity-dependent G1 arrest followed by cell death. Cancer Research, 59(22),
5808–5814.
[3]. Fusco,
N., Sajjadi, E., Venetis, K., Gaudioso, G., Lopez, G., Corti, C., Rocco, E. G.,
Criscitiello, C., Malapelle, U., & Invernizzi, M., 2020, PTEN alterations
and their role in cancer management: Are we making headway on precision
medicine? Genes, 11(7), 719. https://doi.org/10.3390/genes11070719
[4]. Liu,
T., Wang, Y., Wang, Y., & Chan, A. M., 2019, Multifaceted regulation of
PTEN subcellular distributions and biological functions. Cancers, 11(9),
1247. https://doi.org/10.3390/cancers11091247
[5]. Hopkins,
B. D., Hodakoski, C., Barrows, D., Mense, S. M., & Parsons, R. E., 2014,
PTEN function: the long and the short of it. Trends in Biochemical
Sciences, 39(4), 183–190. https://doi.org/10.1016/j.tibs.2014.02.006
[6]. Chen,
C.-Y., Chen, J., He, L., & Stiles, B. L., 2018, PTEN: Tumor suppressor and
metabolic regulator. Frontiers in Endocrinology, 9,
338. https://doi.org/10.3389/fendo.2018.00338
[7]. Luongo,
F., Colonna, F., Calapà, F., Vitale, S., Fiori, M. E., & De Maria, R., 2019,
PTEN tumor-suppressor: The dam of stemness in cancer. Cancers, 11(8),
1076. https://doi.org/10.3390/cancers11081076
[8]. Ngeow,
J., & Eng, C., 2020, PTEN in hereditary and sporadic cancer. Cold
Spring Harbor Perspectives in Medicine, 10(4), a036087. https://doi.org/10.1101/cshperspect.a036087
[9]. Nero,
C., Ciccarone, F., Pietragalla, A., & Scambia, G., 2019, PTEN and
gynecological cancers. Cancers, 11(10), 1458. https://doi.org/10.3390/cancers11101458
[10]. Choi,
S. W., Lee, Y., Shin, K., Koo, H., Kim, D., Sa, J. K., Cho, H. J., Shin, H.-M.,
Lee, S. J., Kim, H., Chung, S., Shin, J., Lee, C., & Nam, D.-H., 2021,
Mutation-specific non-canonical pathway of PTEN as a distinct therapeutic
target for glioblastoma. Cell Death & Disease, 12(4),
374. https://doi.org/10.1038/s41419-021-03657-0
[11]. Wang,
J., Zhang, S., Zhang, J., Zhang, Z., Ma, Q., Fu, W., Chen, X., Zhao, D., Zhao,
M., Di, C., & Xie, X., 2023, A novel PTEN mutant caused by polymorphism in
cis-regulatory elements is involved in chemosensitivity in breast cancer. American
journal of cancer research, 13(1), 86–104.
[12]. Deng,
N., Zhou, H., Fan, H., & Yuan, Y., 2017, Single nucleotide polymorphisms
and cancer susceptibility. Oncotarget, 8(66),
110635–110649. https://doi.org/10.18632/oncotarget.22372
[13]. Ng, P.
C., & Henikoff, S., 2003, SIFT: Predicting amino acid changes that affect
protein function. Nucleic Acids Research, 31(13),
3812–3814. https://doi.org/10.1093/nar/gkg509
[14]. Behairy,
M. Y., Soltan, M. A., Adam, M. S., Refaat, A. M., Ezz, E. M., Albogami, S.,
Fayad, E., Althobaiti, F., Gouda, A. M., Sileem, A. E., Elfaky, M. A., Darwish,
K. M., & Alaa Eldeen, M., 2022, Computational analysis of deleterious SNPs
in NRAS to assess their potential correlation with carcinogenesis. Frontiers
in Genetics, 13, 872845. https://doi.org/10.3389/fgene.2022.872845
[15]. Ajith,
A., & Subbiah, U., 2023, In silico screening of non-synonymous SNPs in
human TUFT1 gene. Journal, Genetic Engineering & Biotechnology, 21(1),
95. https://doi.org/10.1186/s43141-023-00551-4
[16]. Rentzsch,
P., Witten, D., Cooper, G. M., Shendure, J., & Kircher, M., 2019, CADD:
predicting the deleteriousness of variants throughout the human genome. Nucleic
Acids Research, 47(D1), D886–D894. https://doi.org/10.1093/nar/gky1016
[17]. Liu,
X., Wu, C., Li, C., & Boerwinkle, E., 2016, DbNSFP v3.0: A one-stop
database of functional predictions and annotations for human nonsynonymous and
splice-site SNVs. Human Mutation, 37(3), 235–241. https://doi.org/10.1002/humu.22932
[18]. Khan,
I., Ansari, I. A., Singh, P., & Dass J, F. P., 2017, Prediction of
functionally significant single nucleotide polymorphisms in PTEN tumor
suppressor gene: An in-silico approach. Biotechnology and Applied
Biochemistry, 64(5), 657–666. https://doi.org/10.1002/bab.1483
[19]. Naidu,
C. K., & Suneetha, Y., 2016, Prediction and analysis of breast cancer
related deleterious non-synonymous single nucleotide polymorphisms in the PTEN
gene. Asian Pacific Journal of Cancer Prevention: APJCP, 17(4),
2199–2203. https://doi.org/10.7314/apjcp.2016.17.4.2199
[20]. Song,
D.-D., Zhang, Q., Li, J.-H., Hao, R.-M., Ma, Y., Wang, P.-Y., & Xie, S.-Y.,
2017, Single nucleotide polymorphisms rs701848 and rs2735343 in PTEN increases
cancer risks in an Asian population. Oncotarget, 8(56),
96290–96300. https://doi.org/10.18632/oncotarget.22019
[21]. Han,
M., Wu, G., Sun, P., Nie, J., Zhang, J., & Li, Y., 2015, Association of
genetic polymorphisms in PTEN and additional interaction with alcohol
consumption and smoking on colorectal cancer in Chinese population. International
Journal of Clinical and Experimental Medicine, 8(11),
21629–21634
[22]. Andreassen,
K. E., Kristiansen, W., Karlsson, R., Aschim, E. L., Dahl, O., Fosså, S. D.,
Adami, H.-O., Wiklund, F., Haugen, T. B., & Grotmol, T., 2013, Genetic
variation in AKT1, PTEN and the 8q24 locus, and the risk of testicular germ
cell tumor. Human Reproduction (Oxford, England), 28(7),
1995–2002. https://doi.org/10.1093/humrep/det127

